
An Australian Python. Photo by David Clode.
Depending on who you talk to, you may get weird looks for telling someone that you are an R user or a Python user. Both languages have developed significantly over the last few decades and nowadays this language rivalry is unnecessary or simply non-existent.
Aside from the culture war, it is now possible to use one within the others’ environment,
using easily installable packages. Notably, rpy2 and reticulate to use
R within Python and Python within R, respectively.
Try it in Renku
I’ve hosted a sandboxed project on Renku, a platform that I work with on a daily basis. Here you can access an anonymous session with one click: https://renkulab.io/projects/learn-renku/teaching-on-renku/using-r-and-python-together/sessions/new?autostart=1.
Once your session loads up, you will be greeted with RStudio as the default end-point, but don’t worry if this isn’t your preferred editor: we will switch to JupyterLab shortly and you can even use VSCode.
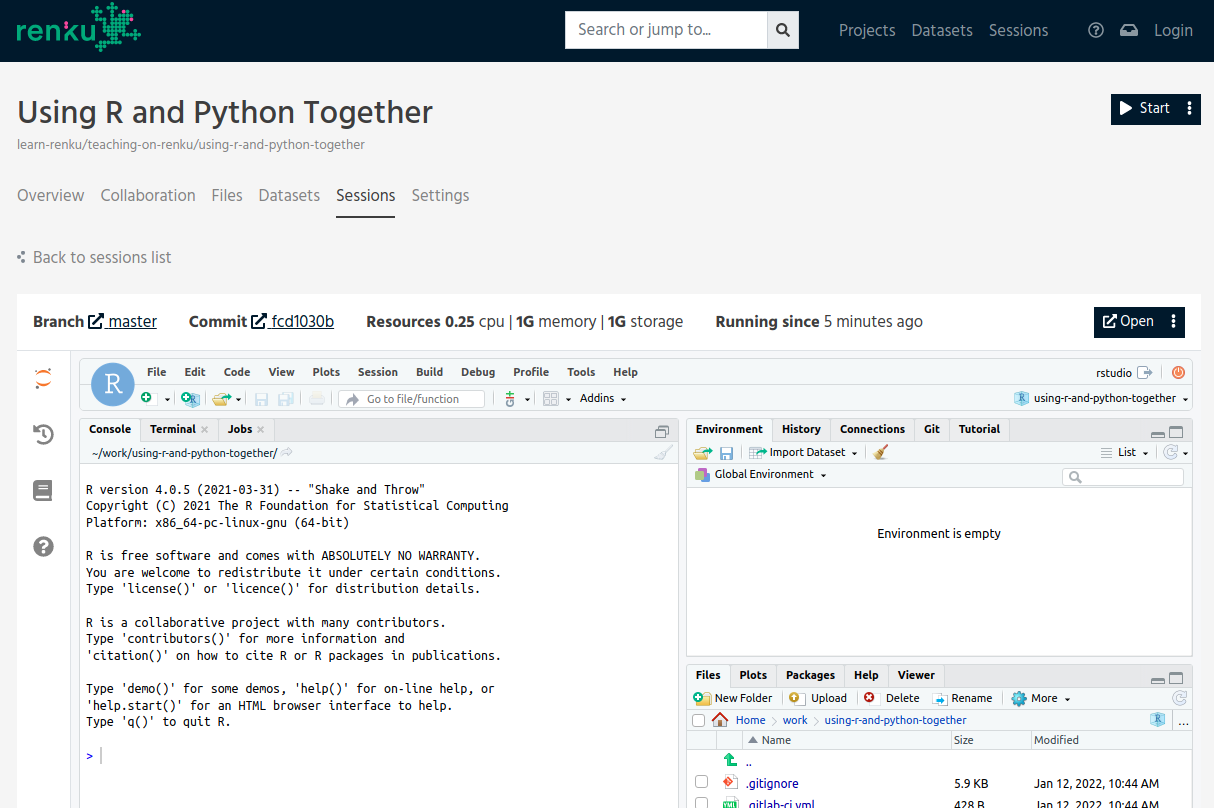
Once your session loads up.
For the purposes of this project, we obviously need both R and Python installed somewhere in order to use them both. We used an RStudio template provided by Renku which actually contains both R and Python.
Python in R
If you open up the ./notebooks/Python_in_R.html
file, you will see how we have called the Python kernel within an R Markdown using Reticulate. You need to point R to where
your Python kernel is. In this case, it is at /opt/conda/bin/python, which is
the default for Renku projects.
```{r load_reticulate}
library(reticulate)
use_python("/opt/conda/bin/python", required = TRUE)
```And that’s essentially it - you can now use Python in your code chunks. For example
```{python pandas}
import pandas as pd
df = pd.DataFrame({'X': [1, 2], 'Y': [3, 4]})
print(df)
```Your Python variables are saved in the special py object, so you can access them
easily!
```{r access_py}
py$df
```which outputs:
## X Y
## 1 1 3
## 2 2 4To JupyterLab
As I mentioned before, this particular sandbox comes with both RStudio and JupyterLab! You will have to open the session in a new tab and then change the end-point to lab:
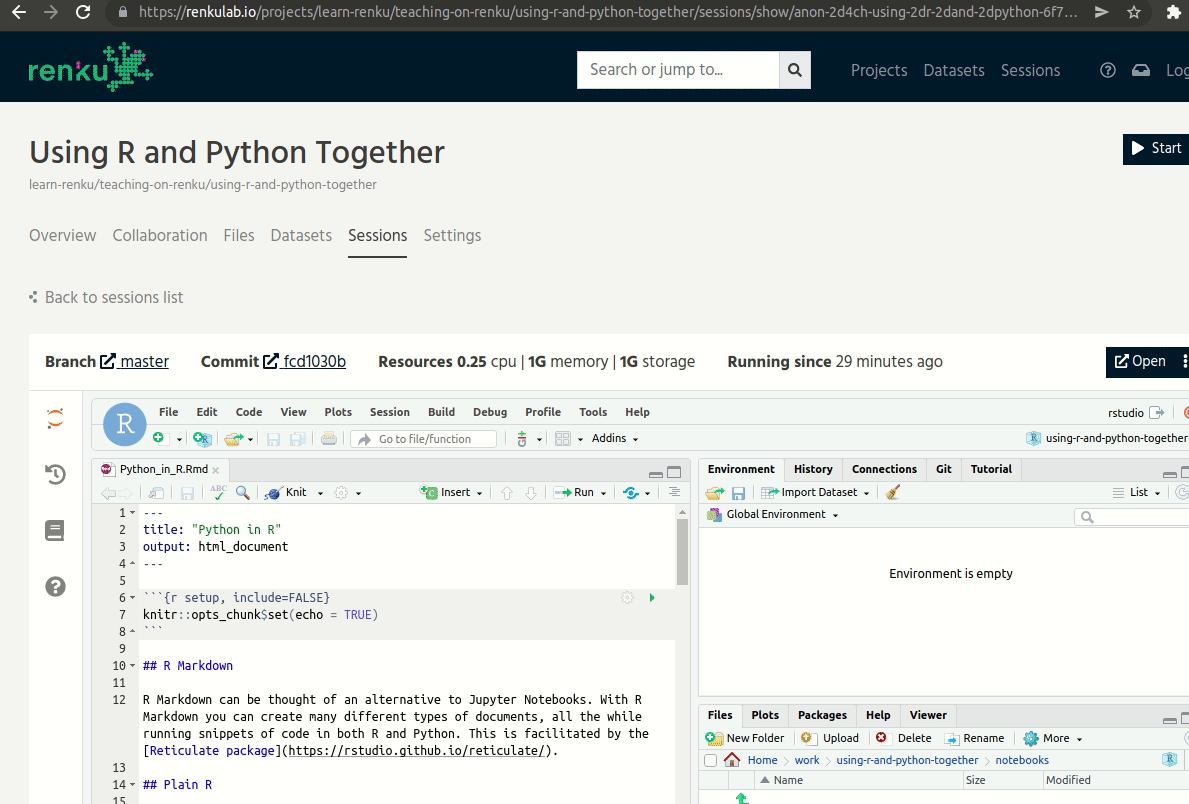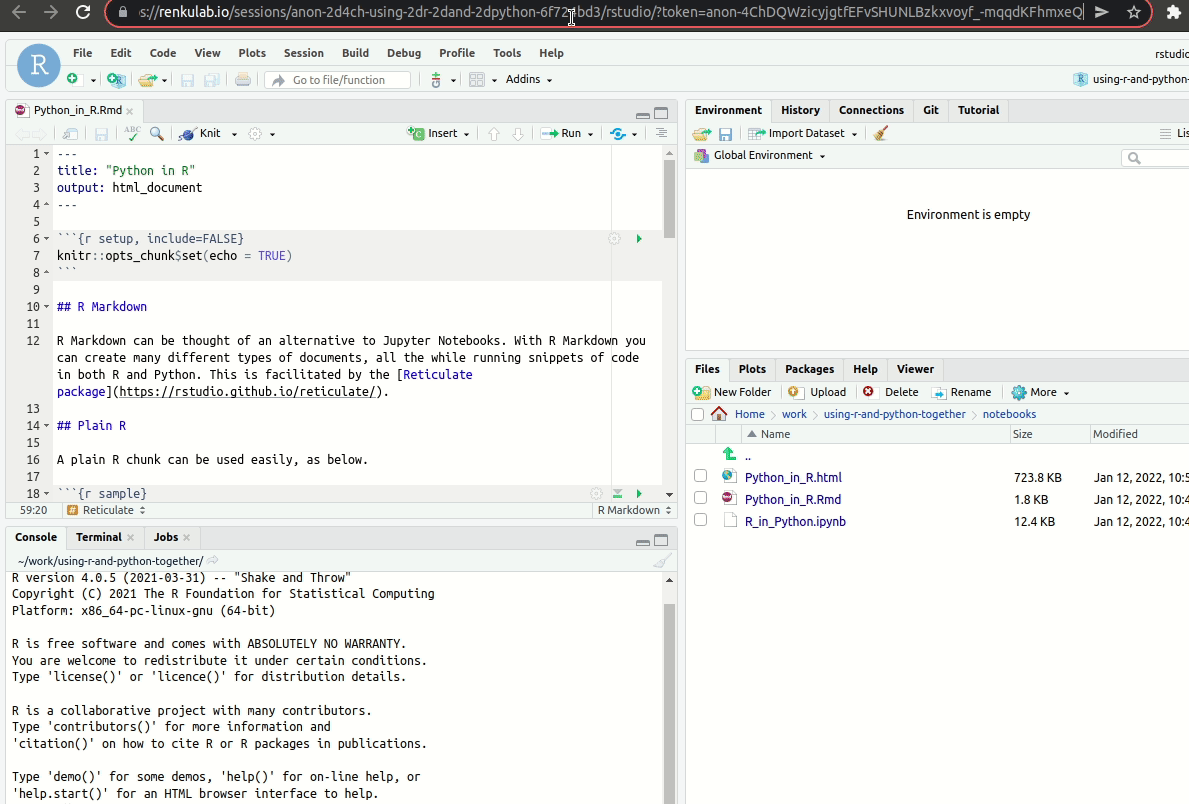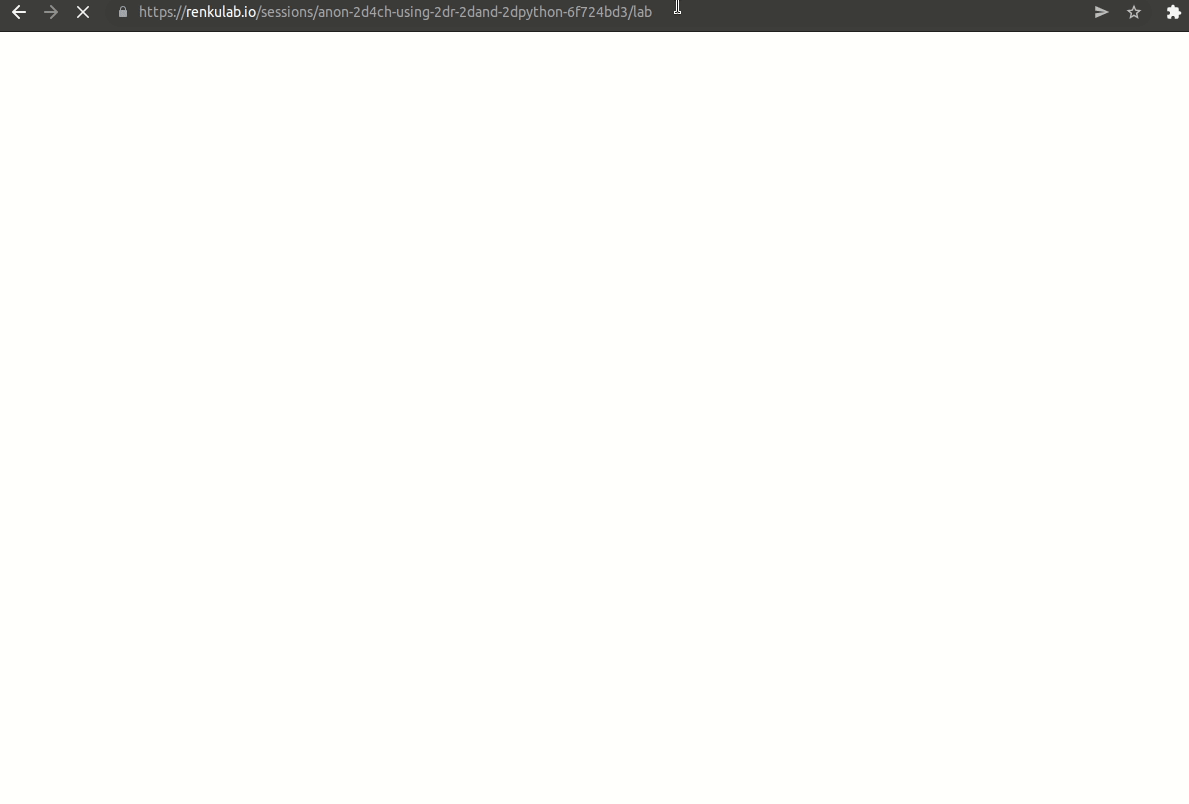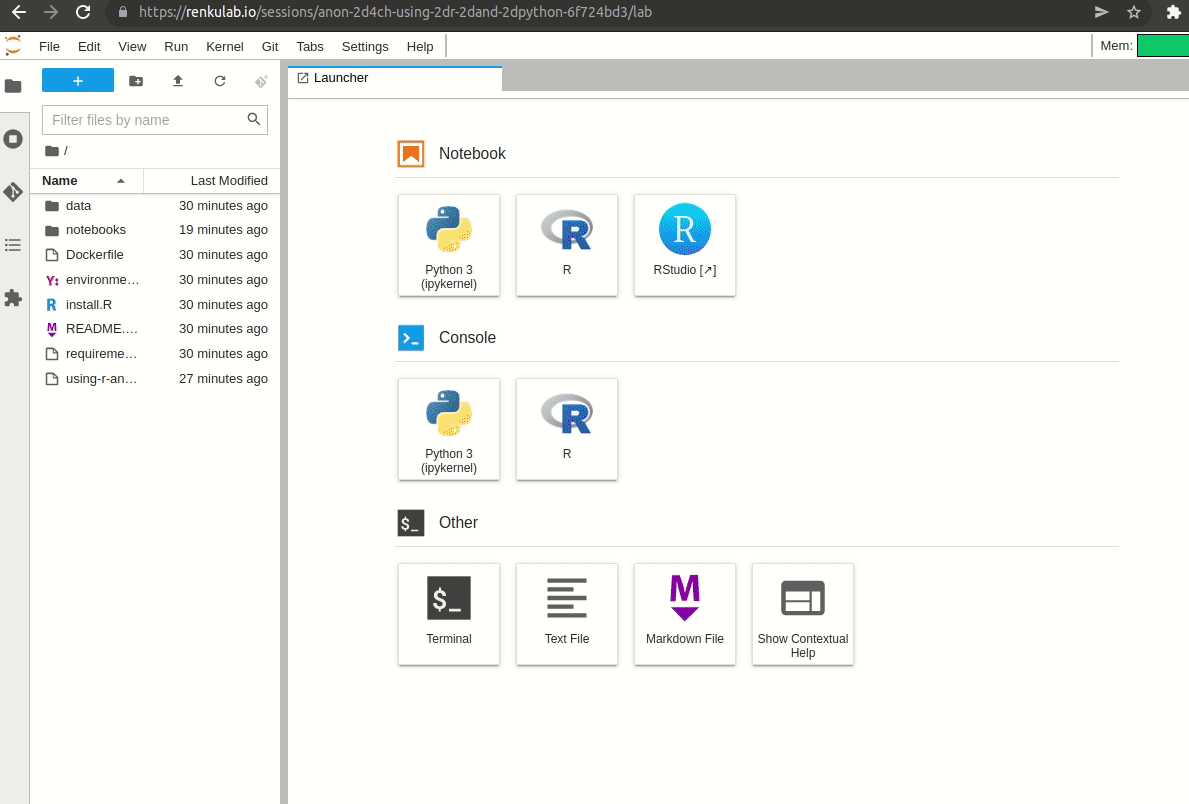
To JupyterLab from RStudio.
Python in R
This is even easier than Python in R. The rpy2 package
gives us Jupyter magic in the form of %R for in-line commands or %%R for an
entire cell. All you have to do is load the extension:
%load_ext rpy2.ipythonA multi-line cell is given as an example in the ./notebooks/R_in_Python.ipynb:
%%R
regression <- lm(mtcars$mpg ~ mtcars$carb)
summary(regression)Interfacing numpy and R matrices is also easy, as you will find further down in that notebook.
Safe-keeping
If you want to save a copy of this sandboxed environment for later use, fork the following project into your own namespace: https://renkulab.io/projects/learn-renku/teaching-on-renku/using-r-and-python-together. You will need a Renku account, but you can use GitHub to login if you already have this.
Truce?
Both R and Python are widely popular and there is no reason to eschew one in favour of another. They are capable of many things and now it is easy to lean into the other if need.
